tRex is a database of tRNA-derived short fragments (tRFs) and tRNAs in Arabidopsis thaliana
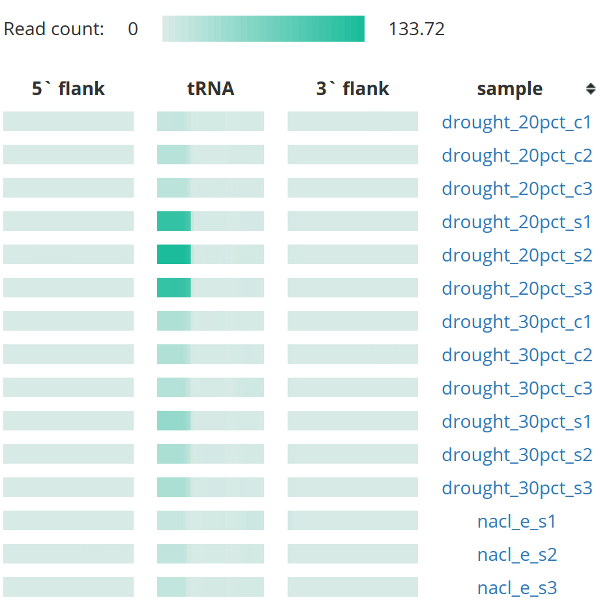
tRex employs different options for data exploration, including per sample or per tRNA browsing, overview of simplified expression patterns across different samples or tRNAs and full expression profiles at read-level resolution. Each profile view is enabled with dynamic rescaling for different methods of normalization and connected with other pertinent information in an easy to navigate manner.
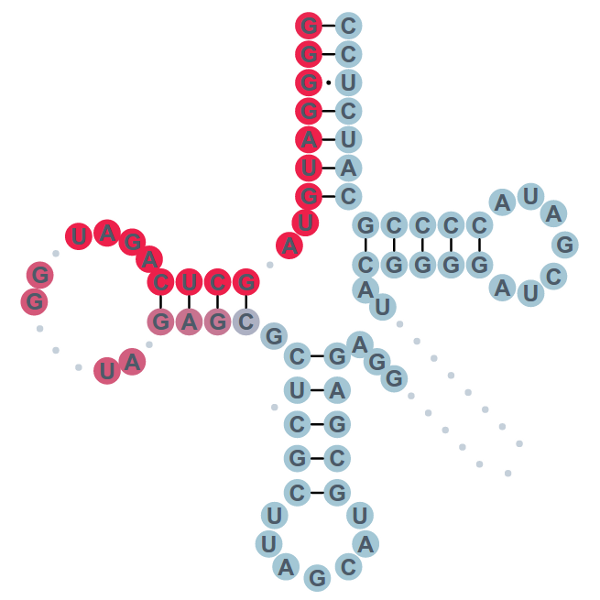
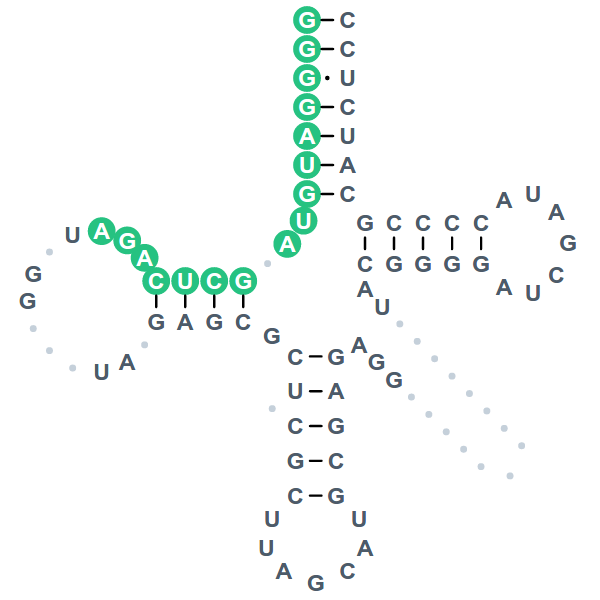
Annotated tRNAs are displayed as a sequence, cloverleaf or MFE secondary structure in a two-panel window for ease of comparison. When viewed per sample, structural profiles include information about read coverage and predicted types and positions of modified nucleotides.
Information about tRNA-derived fragments includes a table of all tRNAs to which tRF is mapping, its localization displayed on a canonical structure, any of its predicted mRNA or transposable element targets as well as all samples in which the fragment is detected.