Help about
There are 3 ways to access ribosomal 5S RNA records within the 5SrRNA database:
To find individual 5S rRNA records, you can start typing either a particular record identifier (e.g., E00204) or species (e.g., Homo sapiens) or a higher taxon name (e.g., Mammals).
If you don't remember the scientific name of a taxon, you can use its common name (e.g. human, man) or synonym.

Taxonomy Browser lets you retrieve individual sequences from a particular organism as well as sets of sequences from related species. You can select either the single species (last node in the tree) or any intermediate junction to retrieve records for all species included within a particular taxonomic branch.
The classification of organisms used by the browser is based on the NCBI Taxonomy Database.
The numbers shown along with taxon name indicate the number of species - for example 5282 - and records associated with each node - for example: 7291.
Use BLAST tool if you want to find homologous sequence(s) within 5SrRNAdb with your custom query sequence
Just paste your sequence in a form box and click the "BLAST" button.
You can also specify BLAST parameters such as: BLAST algorithm, E-value cut-off, identity and score cut-offs using the circle widgets.
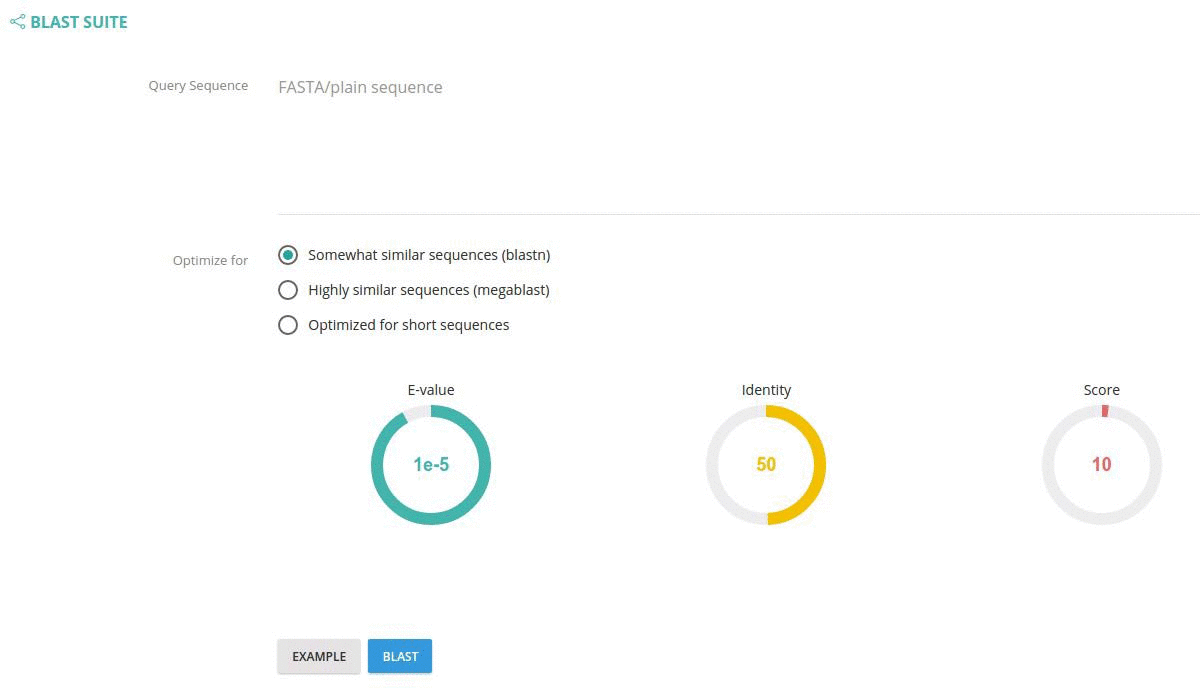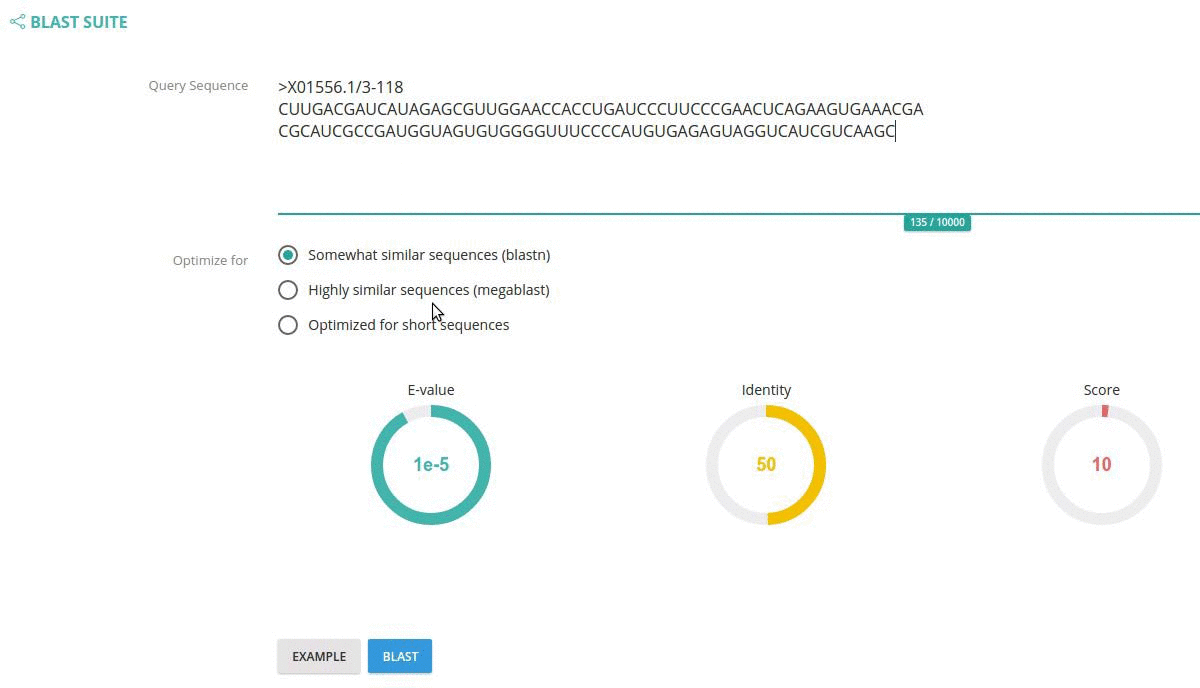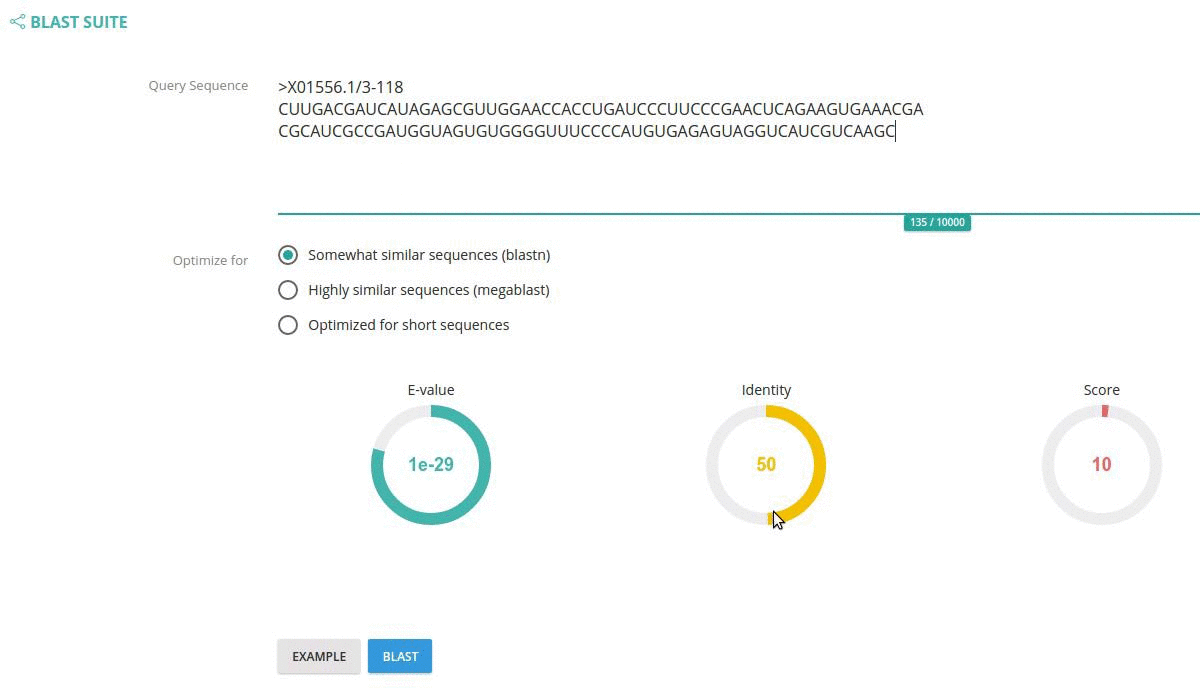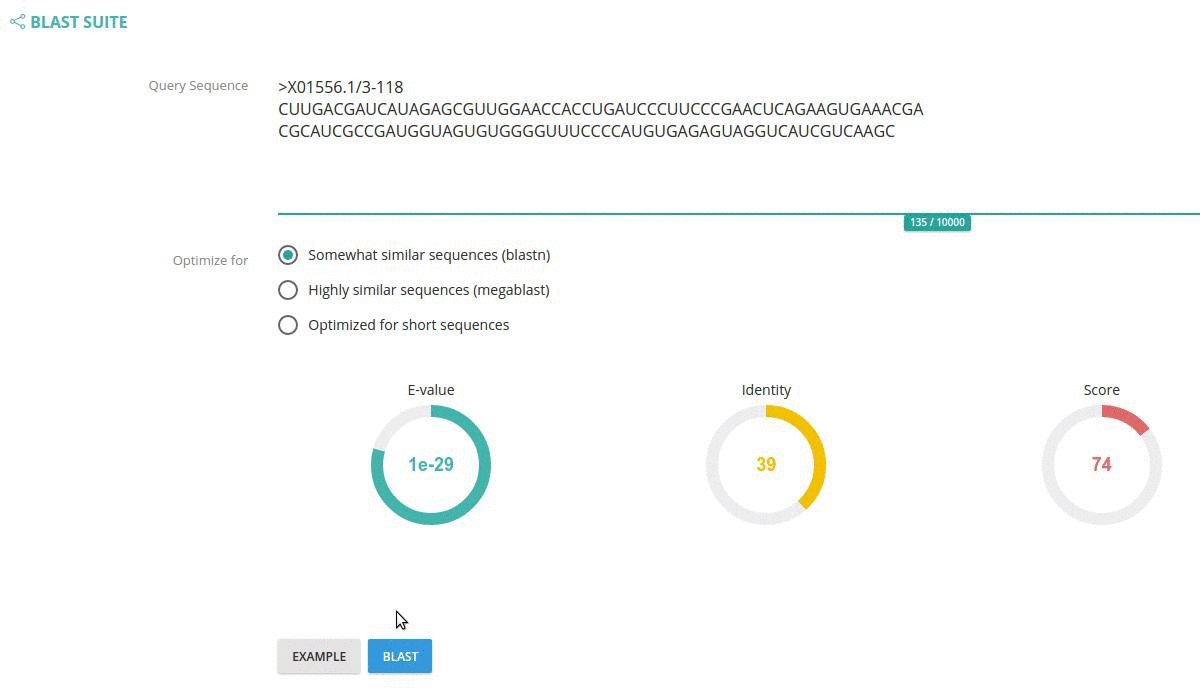
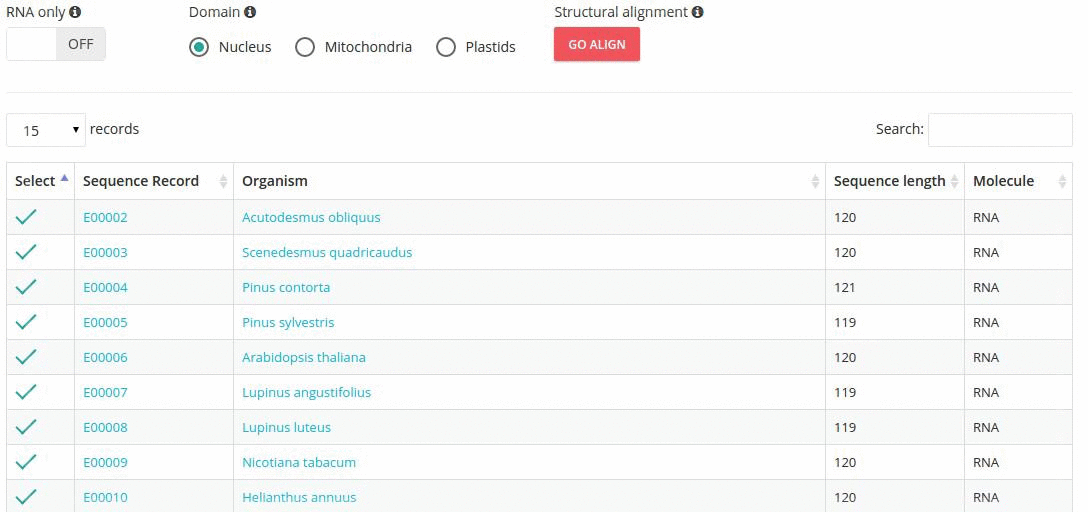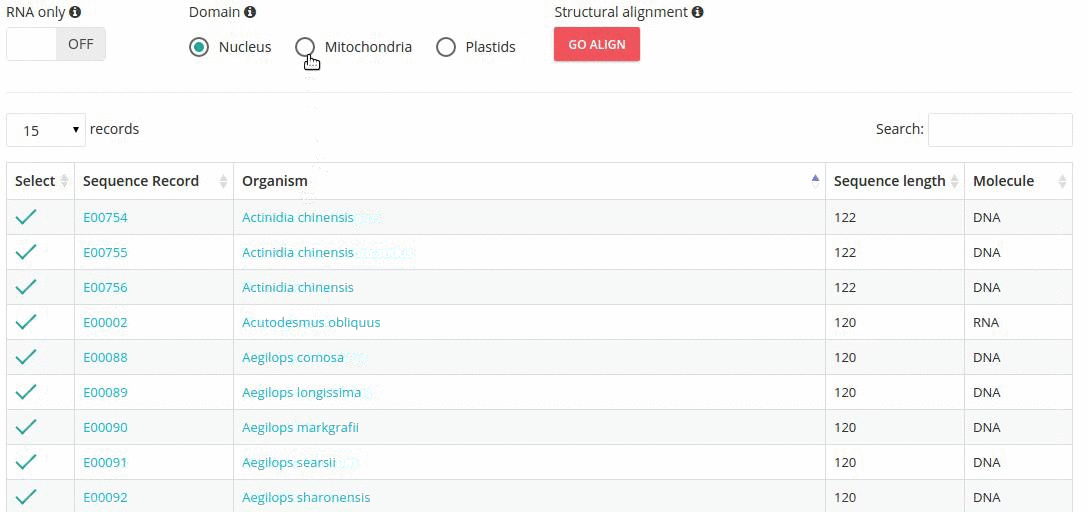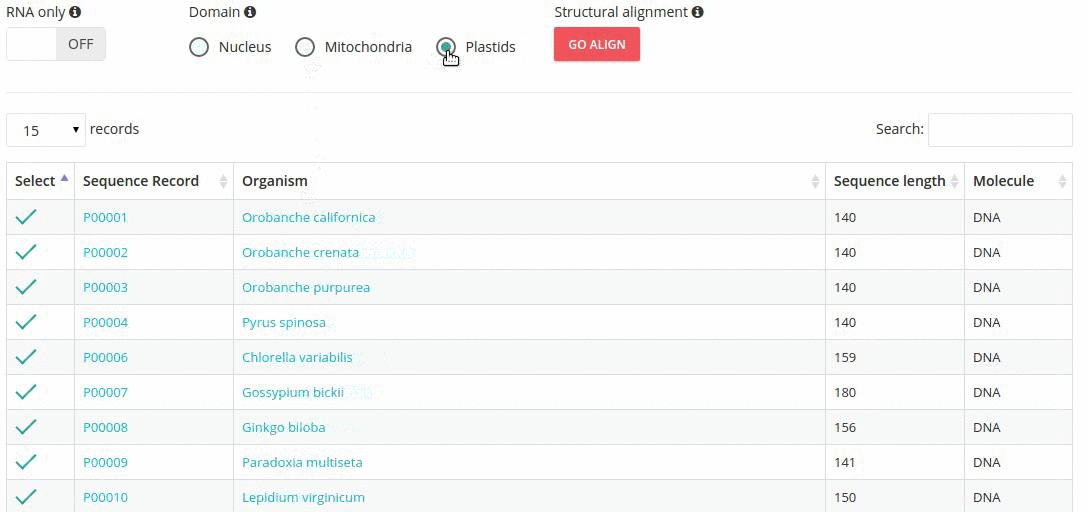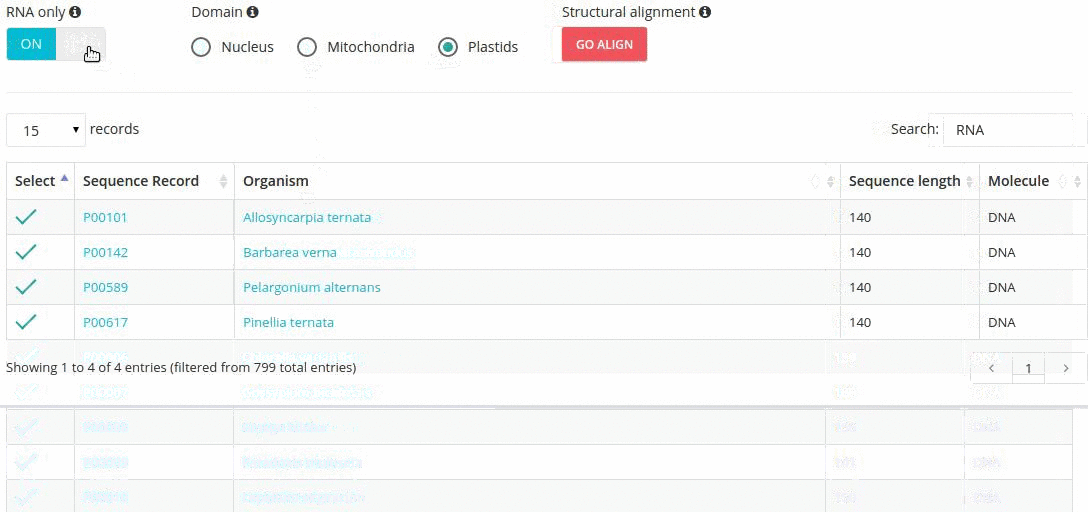
Records matching a query (Text search, Taxonomy Browser or BLAST) are shown in a table providing record identifier, organism name, sequence length and the type of sequenced molecule (RNA or DNA).
Turning the 'RNA only' option, results in search limited to records containing sequences that were obtained by direct (enzymatic or chemical) RNA sequencing methods. Specifing the domain option (i.e., nucleus, plastids, mitochondria) further restricts the range of resulting records.
In addition, the results table provides ways of filtering results such as:
- search-based text filtering
- column sorting
- customized pagination
Each individual database record represents a unique 5S rRNA sequence identified in a particular species. In addition to the source organism name and the nucleotide sequence each record contains information on the phylogenetic position of the organism linked to the taxonomy resources at the NCBI.
The primary sources of the sequence are listed as links to the original GenBank records.
Each record contains information on the secondary structure that is displayed as a diagram. The structure diagrams show nucleotide sequences in the context of the general secondary structure models containing all positions in the alignments.
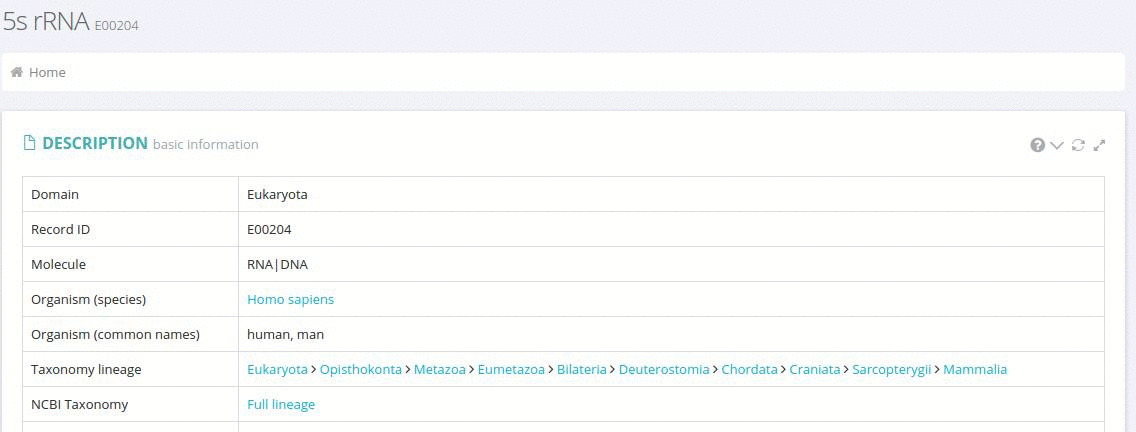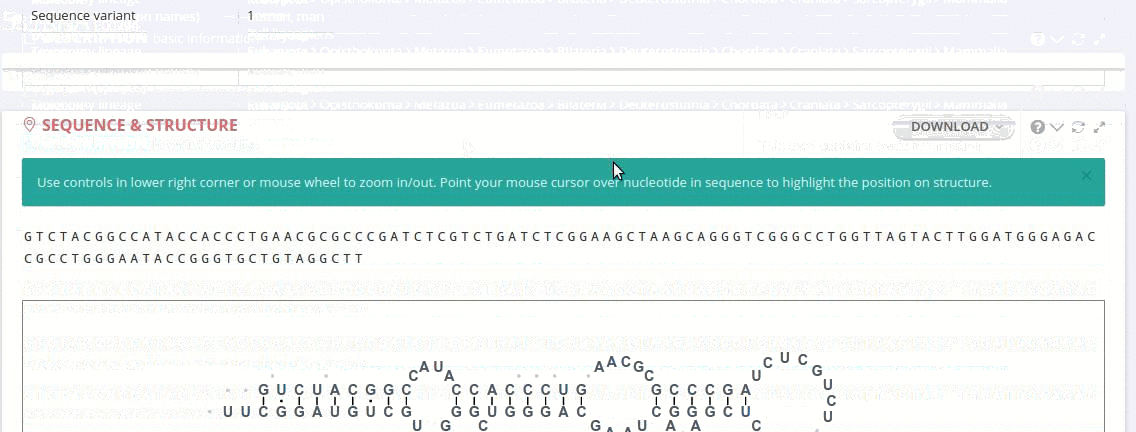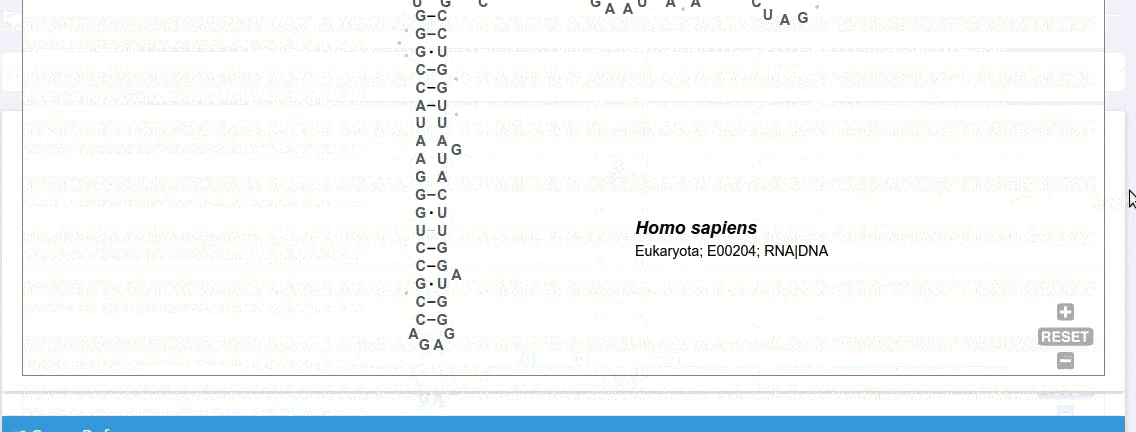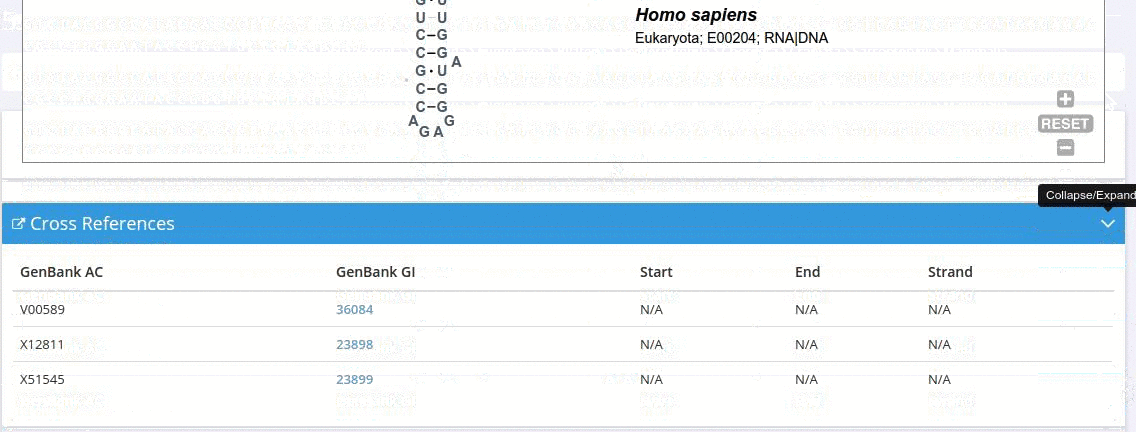
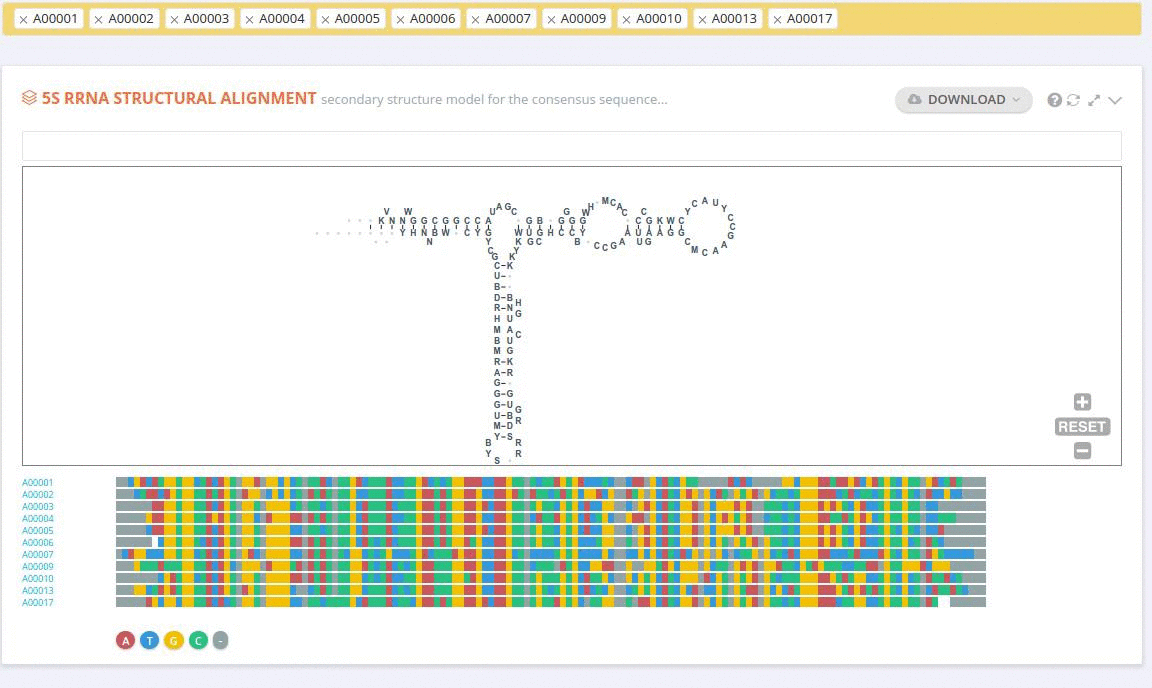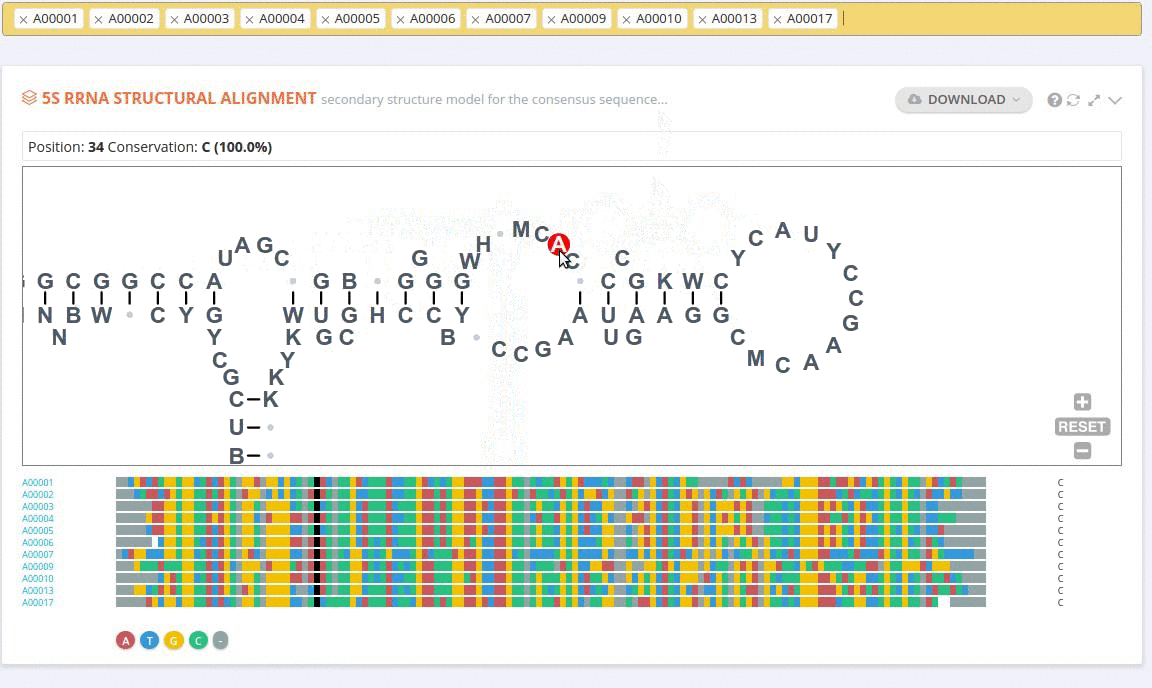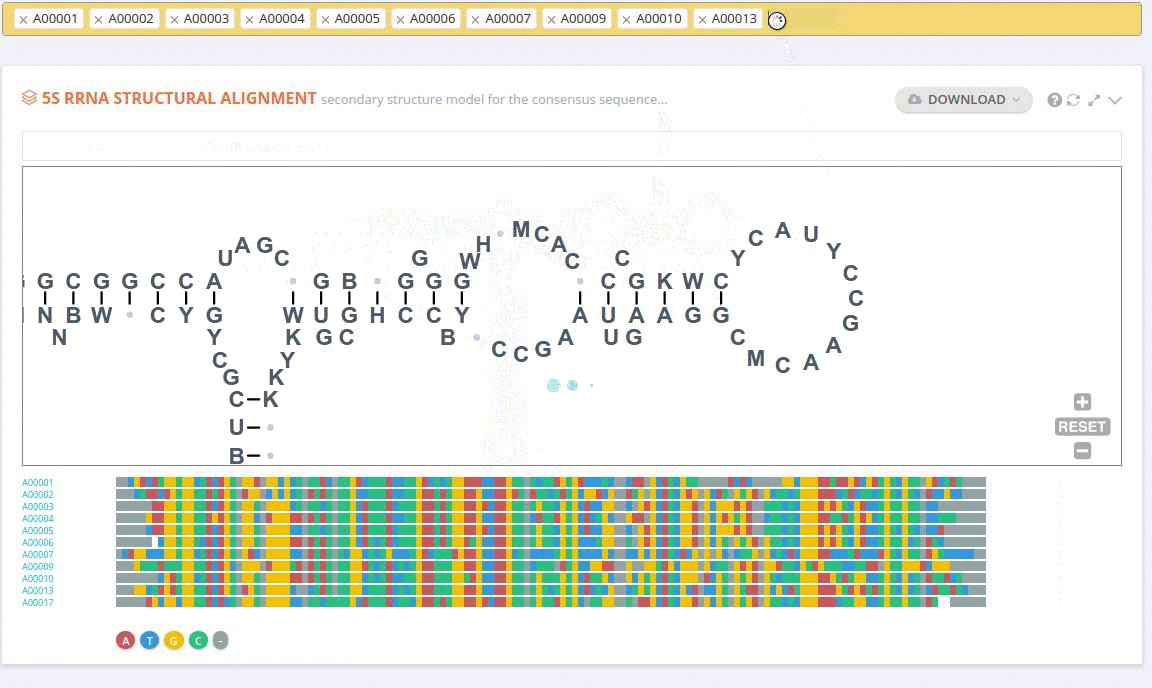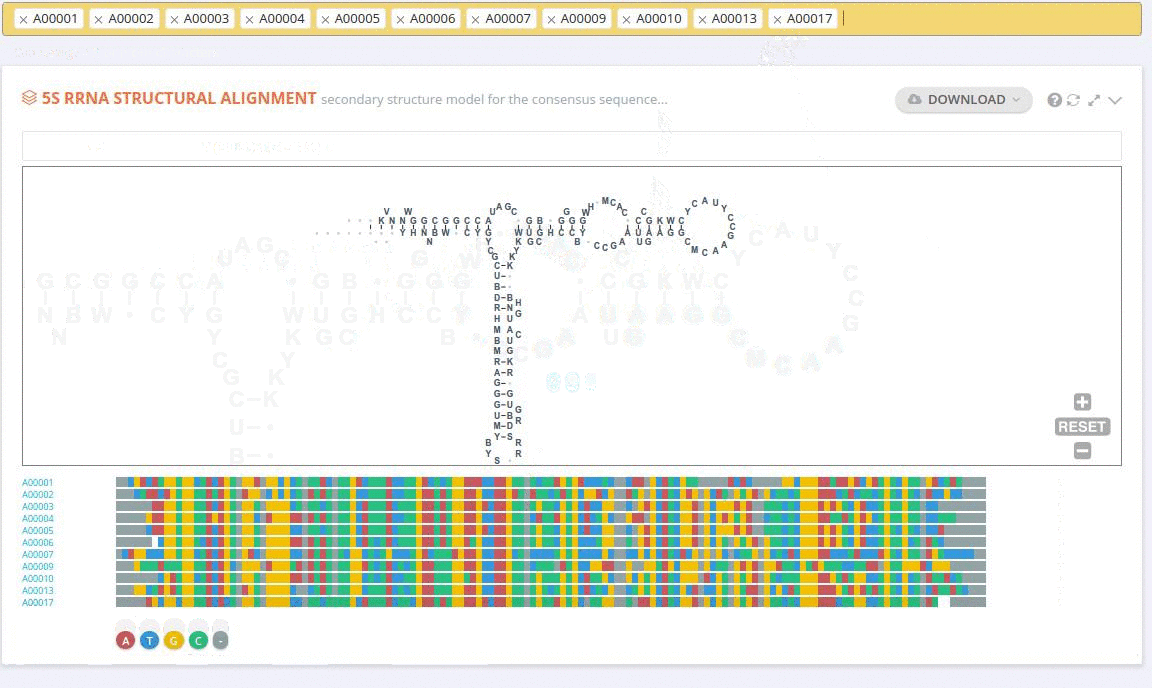
The ‘Alignment’ framework contains a secondary structure model representing the consensus sequence derived from the current alignment.
The secondary structure diagrams are interactive - the nucleotides or base pairs statistics are displayed when pointing at a nucleotide or base pair symbol. To customize the content of the alignment, you can add and/or remove sequences from the current view using the panel listing records or by providing the record identifier. The new nucleotide statistics and the secondary structure model are dynamically recalculated to match the current set of sequences in the alignment.
The custom alignment can also be built from scratch by adding together results of subsequent search queries.
The user interface of the 5S rRNA database has several user-oriented solutions enhancing the efficiency of the data mining experience. All browsing and searching results are now presented as separate, clearly named windows with both graphical elements providing contextual clues as well as mouse-over tooltips.
Users can adjust the amount of information that is present on each page, as any window can be dynamically wrapped up or expanded.
All data that have been used to create the database can be downloaded from interactive windows